paired end sequencing insert size
It also provides functions to calculate metrics for validating library. Multiplex sequencing of paired-end ditags MS-PET.
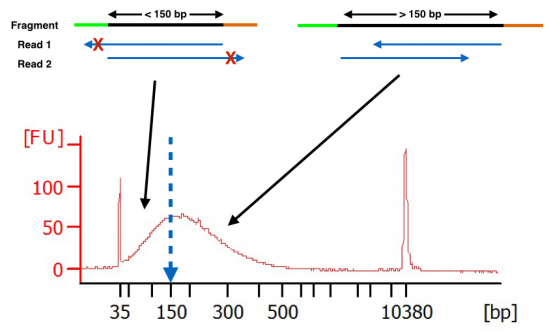
How Short Inserts Affect Sequencing Performance
Rob Edwards from San Diego State University describes how Illumina paired-end sequencing works.

. Download scientific diagram Paired-end PE sequencing and insert size IS filtering to increase sensitivity. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a. Source Name CommentENA_SAMPLE CommentBioSD_SAMPLE Characteristicsorganism Characteristicscell line Characteristicsprogenitor cell type Characteristicscell type Characteristicsimmunophenotype Material Type Protocol REF Protocol REF Protocol REF Protocol REF Extract Name CommentLIBRARY_LAYOUT CommentLIBRARY_SELECTION.
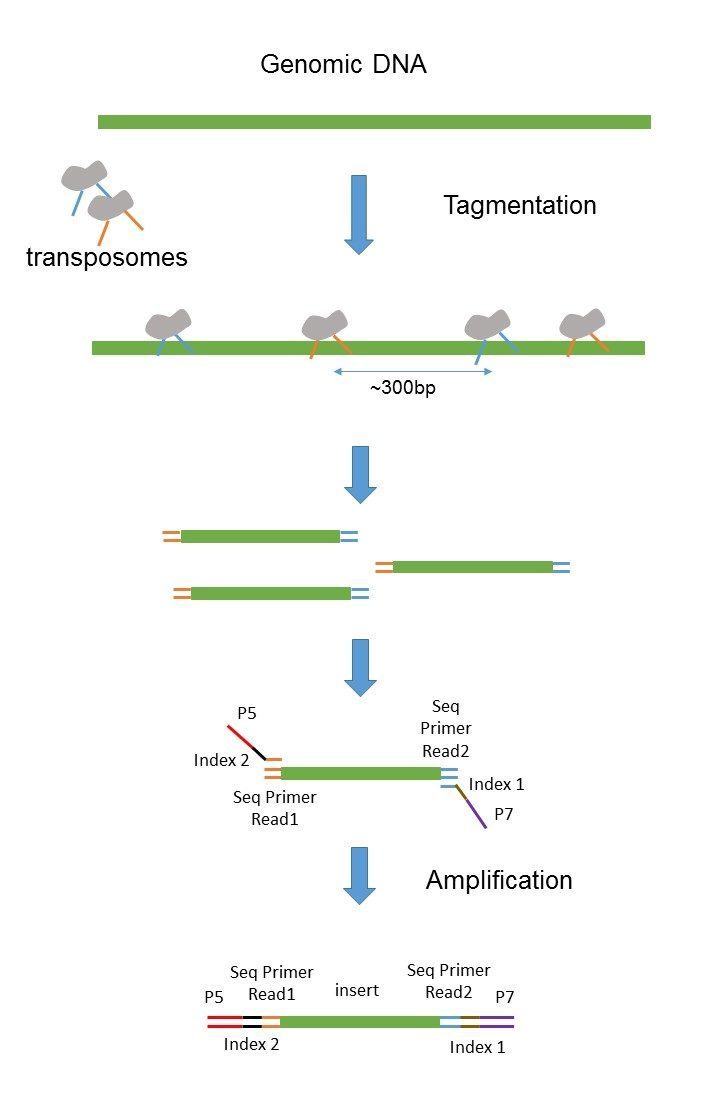
Transpososomes are used to fragment DNA to be sequenced and add. A Sample-wise IS histograms of liquid biopsies LBs. Requires the same amount.
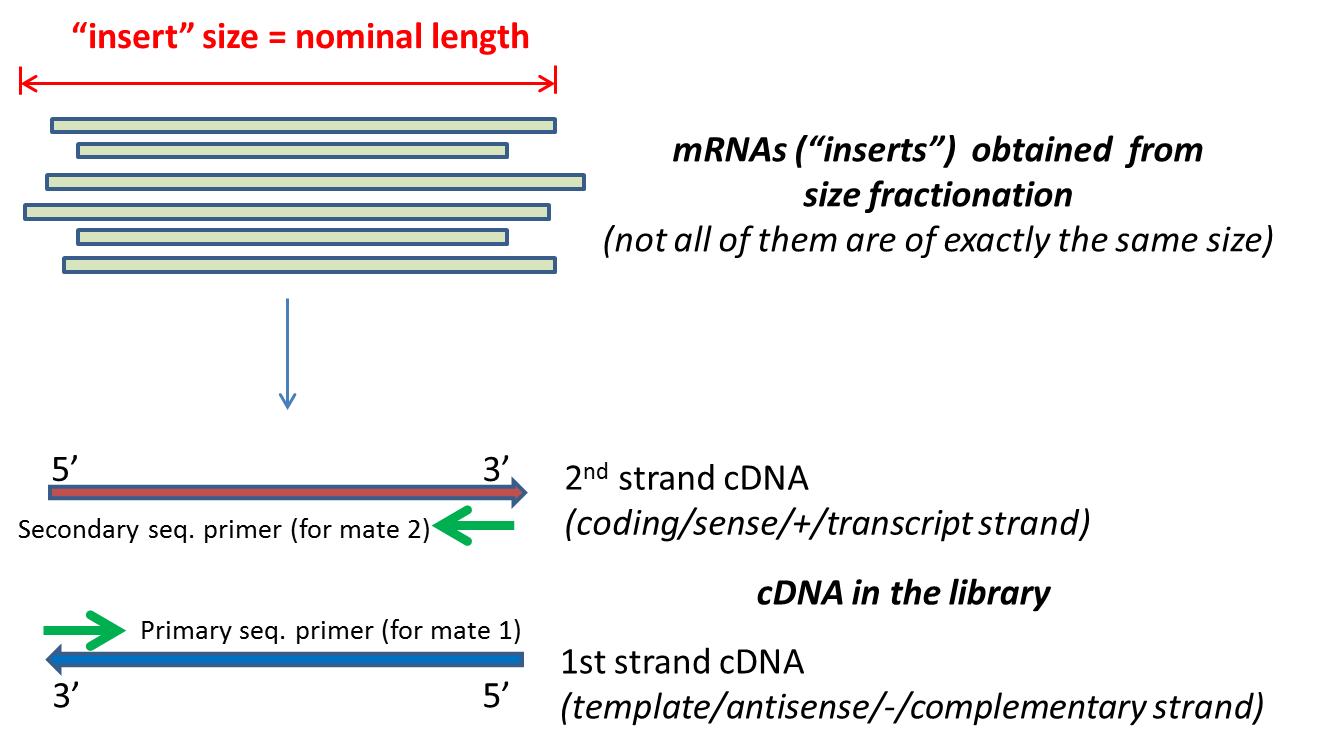
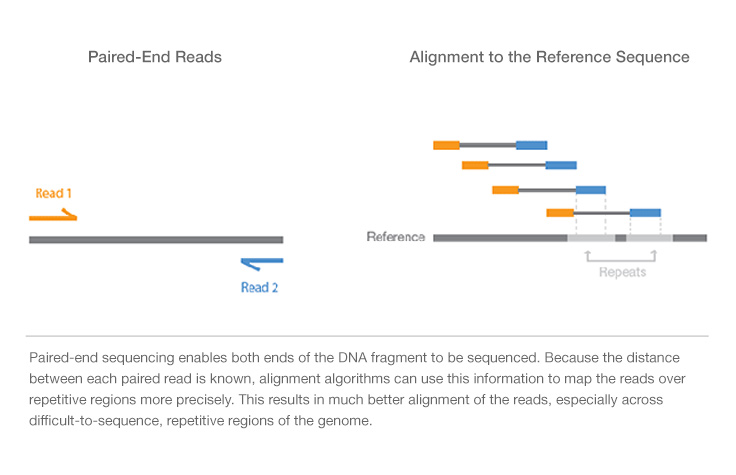
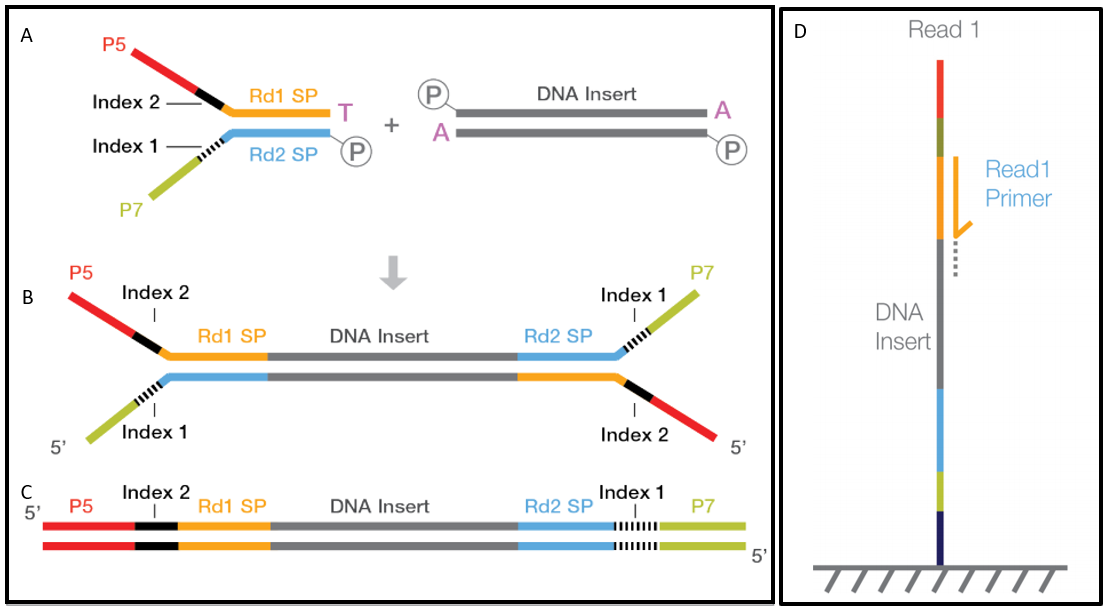
A good choice for read length is closely tied to the insert size of the sequencing library ie how long the individual DNA fragments are that are sequenced. In an Illumina sequencing run either single-end sequencing SE or paired-end sequencing PE can be used. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece.
The Illumina NexteraXT transposon protocol is a cost effective way to generate paired end libraries. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle. This size depends on the library.
While the underlying principles between PE and MP reads. Transpososomes are used to fragment DNA to be sequenced and add adapter. A strategy for the ultra-high-throughput analysis of transcriptomes and genomes.
Department of Energy Office of Scientific and Technical Information. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert. 1Create reference transcriptome index for BowTie or BWA.
As we know by now and the insert size limitation becomes an issue when you want paired-end reads with a higher inner distance with fragments longer than 800 bp. These attributes support sequencing of complex genomes and comprehensive characterization of the widest range of structural variants. The Illumina NexteraXT transposon protocol is a cost effective way to generate paired end libraries.
Paired end sequencing refers to the fact that the fragments sequenced were sequenced from both ends and not just the one as was true for first generation sequencing. Paired-end insert size ranges. In any case DNA is chopped into small fragments ligated with.
B Histology-wise mean IS. Simple workflow allows generation of unique ranges of insert sizes. Paired-End Library Insert Sizes Picard The Picard citation library is used in SeqSphere to process SAMBAM files.
The reads have a length of typically 50 - 300 bp. The sequencing starts at Read 1 Adapter mate 1 and ends with the sequencing from Read 2 Adapter mate 2. 2Align paired-end RNA-Seq data to the reference transcripts using minimum insert-length as zero and.
07-31-2011 1158 PM. The fragment size which you need to select for during a gel purification for example would be the insert size length of both adapters around. In DNA sequencing lingo the words paired-end PE and mate-pair MP are frequently used interchangeably.
Paired End Sequencing France Genomique

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram

What Is Mate Pair Sequencing For

Interpreting Color By Insert Size Integrative Genomics Viewer

What Is Mate Pair Sequencing For

Pdf Assessment Of Insert Sizes And Adapter Content In Fastq Data From Nexteraxt Libraries

Library Insert Size Libraries Were Constructed Using The Standard A Download Scientific Diagram

Interpreting Color By Insert Size Integrative Genomics Viewer

Definition Of Essential Terms A Definition Of Fragment Read And Download Scientific Diagram

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram
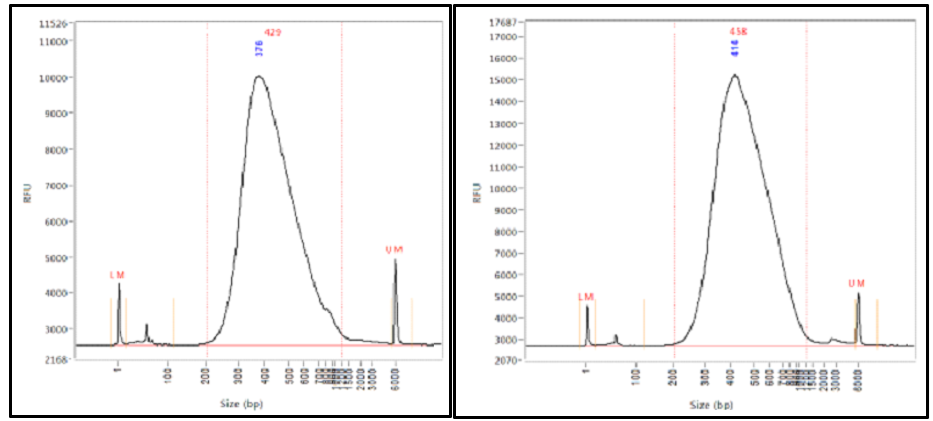
What Happens When Hardware Advances Faster Than Molecular Protocols Cofactor Genomics
How Short Inserts Affect Sequencing Performance

What Are Paired End Reads The Sequencing Center
What Is Paired End 150 Omega Bioservices

What Are Paired End Reads The Sequencing Center

Mp Insert Size Distribution And Library Complexity A Insert Size Download Scientific Diagram
What Read Lengths Are Produced By Modern Illumina Sequencers